ViralEntry™ Transduction Enhancer (100X)
|
Cat. No.
|
G515 |
|---|---|
| Unit |
1.0 ml
|
| Cat. No. | G515 | ||
| Name | ViralEntry™ Transduction Enhancer (100X) | ||
| Unit | 1.0 ml | ||
| Description | (Equivalent to Cat. No. G698, G514, G511, G512, G513)
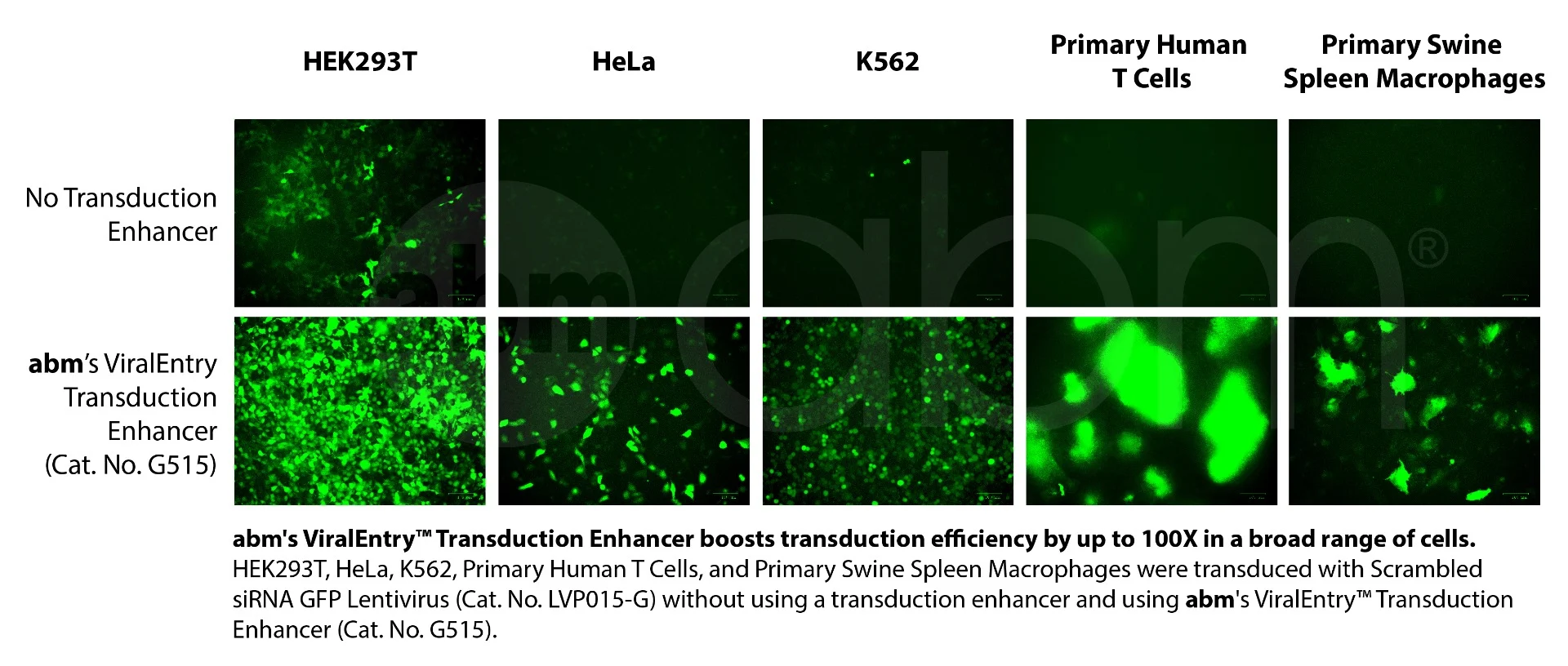
Boost transduction by over 10X in a variety of cell types!As an industry leader in recombinant viral vectors (lentivirus, adenovirus, and AAV), abm has 15+ years of experience in how to enhance virus vector transduction efficiency. Over the years, we have developed multiple formulations of viral enhancing agents useful for this purpose. A remarkable breakthrough by our top scientists in 2020 is the development of the most efficient viral enhancing agent, ViralEntry™, that is not only effective in enhancing lentivirus transduction efficiency significantly, but also increases transduction efficiency in diverse cell types including primary T lymphocytes by over 10X. This is superior to any similar product on the market. With this incredible reagent, you will be able to transduce any cell types you have. We guarantee that this is the most effective viral transduction reagent in the industry and will gladly refund you if you find better transduction enhancers than our ViralEntry™ formulation. Use it and you will enjoy it! Product Features
|
||
| Storage Condition | Store at 4°C. |
What are the correct concentration units for each recombinant viral particle?
For lentiviruses and retroviruses, they are measured in CFU/ml (colony-forming units per millilitre). Transduction with lentiviruses and retroviruses can cause the formation of colonies, which can be quantified for concentration. For AAV the titer is measured as genome copies per mL (GC/mL). Adenoviruses are measured as PFU/ml (plaque-forming units per millilitre). Transduction with adenoviruses will kill packaging cells, forming plaques in the process for quantification. The concentration for all three types of viruses can also be classified as IU/ml (Infectious Units/ml). Ultimately, the units refers to the viral particles and different units reflect the different assays involved.
What do I use to check if my cells were successfully immortalized by the SV40 agent?
We have an SV40 T antibody that can be used for the western blot analysis. The catalog number is G202. Otherwise, a qPCR primer can be designed on the SV40 gene for qPCR analysis. The sequence can be found in the link below: http://www.abmgood.com/pLenti%20SV40-Vector-Location-Map.html
What are the primers to use for SV40 identification?
SV40 Forward Primer Sequence 5’ ACTGAGGGGCCTGAAATGA SV40 Reverse Primer Sequence 5’ GACTCAGGGCATGAAACAGG These are qPCR primers and the band size is 61 bp.
What advantages / disadvantages exist between the Lenti-SV40, -SV40T, and SV40T+t vectors?
There are simply differences in the content of all vectors due to customer demand for variety. Lenti-SV40 will contain the whole SV40 gene, -SV40T, the large T Antigen only, and -SV40T&t the large and small T antigens only. It is up to the end user to decide which vectors will best suit their project, however we have successfully used Lenti-SV40 (whole gene) in a wide range of immortalization projects.
What is the accession number for the SV40?
The SV40 covers the entire genome and the accession number is J02400.1. You can use this information to design primers for conventional PCR as well.
How long after transduction can the infection efficiency be observed?
You can observe transduction efficiency from 48 hours up to 5 days after infection.
What are the primers to use for SV40T and SV40T tsA58 detection?
PCR primers: SV40T Forward Primer Sequence 5’ AGCCTGTAGAACCAAACATT 3' SV40T Reverse Primer Sequence 5’ CTGCTGACTCTCAACATTCT 3' The two primers should amplify the region between 3677-4468bp, giving a 792bp fragment.
What is the sequence of the SV40 large T antigen?
This information can be accessed on this page by clicking on "pLenti-SV40-T" under vector map. The Large T antigen is at position 5079-5927.
For G221 and LV620, what does the 'V12' in RasV12 mean?
The V12 means that amino acid # 12 is mutated from a Valine to a Glycine. Other than that, the sequence matches the coding region of HRAS perfectly (NM_005343).
Where is the SV40T tsA58 gene sequence?
The SV40T tsA58 gene is located between 3138-5264bp, with the Alanine-to-Valine mutation at amino acid 438.
This product has no review yet.
Controls and Related Product:


