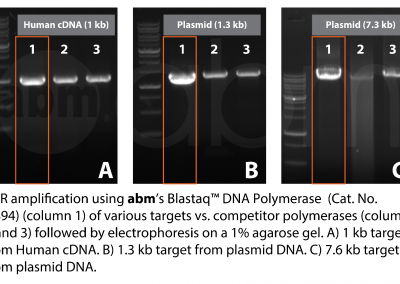
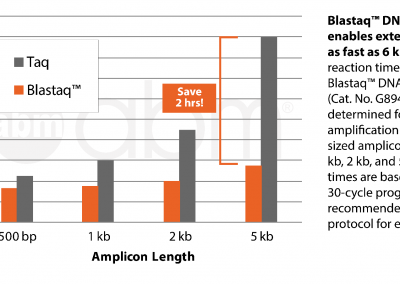
Specifications
| SKU | G894 | |
| Unit quantity | 2000 U | |
| Description | Streamline your PCR with abm’s Blastaq™ DNA Polymerase (5 U/µl). abm’s Blastaq™ DNA Polymerase (5 U/µl) is a next generation Taq Polymerase strategically-engineered to have incredibly fast extension rates and very robust performance. Paired with its uniquely-formulated buffer and specialized reaction conditions this polymerase will provide increased processivity, yields and sensitivity all while reducing total reaction times by 40–70% compared to wild-type Taq DNA polymerase. | |
| Component | 2000 U | |
| Blastaq™ DNA Polymerase (5U/µl) | 400 μl | |
| 5X Blastaq™ Buffer* | 8 ml | |
| 25 mM MgSO4 | 1 ml | |
| *contains Mg2+ | ||
| CHARACTERISTIC | BlasTaq™ |
| Proofreading | No |
| Fidelity (vs. Native Taq) | 1X |
| Specificity | ●●● |
| Extension Speed | 15 sec/kb |
| Target Length | 10-15 kb |
| Suitable for TA cloning | Yes |
| Format | Enzyme (G894) |
| MasterMix (G895) | |
| Special Feature | Fast and Robust |
dNTP Mix
| 제품 번호 | 품 명 및 규 격 | ||
| G050 | dNTP Set 4 x 250 µl (100 mM each) | ||
| G010 | dNTP Mix 250 µl, 10 mM each | ||
| G128 | dNTP Mix 500 µl, 10 mM each | ||
| G129 | dNTP Mix 1.0 ml, 10 mM each | ||
Documents
Supporting Protocol
Other
FAQs
References
- Chung, G et al. “CHL-1 provides an essential function affecting cell proliferation and chromosome stability in Caenorhabditis elegans” DNA Repair (Amst.) 10 (11):1174- 1182 (2011). DOI: 10.1016/j.dnarep.2011.09.011. PubMed: 21968058. Application: PCR.
- Jones, MR et al. “The atm-1 gene is required for genome stability in Caenorhabditis elegans” Mol. Genet. Genomics 287 (4):325-335 (2012). DOI: 10.1007/s00438-012-0681-0. PubMed: 22350747. Application: PCR.
- Ahn, CH et al. “Bacterial biofilm-community selection during autohydrogenotrophic reduction of nitrate and perchlorate in ion-exchange brine” Appl. Microbiol. Biotechnol. 81 (6):1169-1177 (2009). DOI: 10.1007/s00253-008-1797-3. PubMed: 19066883. Application: PCR.
- Bhagwat, B et al. “An in vivo transient expression system can be applied for rapid and effective selection of artificial microRNA constructs for plant stable genetic transformation” J Genet Genomics 40 (5):261-270 (2013). DOI: 10.1016/j.jgg.2013.03.012. PubMed: 23706301. Application: PCR.
- Chung, G et al. “CHL-1 provides an essential function affecting cell proliferation and chromosome stability in Caenorhabditis elegans” DNA Repair (Amst.) 10 (11):1174-1182 (2011). DOI: 10.1016/j.dnarep.2011.09.011. PubMed: 21968058. Application: PCR.
- Lee, G et al. “Inhibition of in vitro tumor cell growth by RP215 monoclonal antibody and antibodies raised against its anti-idiotype antibodies” Cancer Immunol. Immunother. 59 (9):1347-1356 (2010). DOI: 10.1007/s00262-010-0864-7. PubMed: 20473495. Application: PCR.
- Lee, G et al. “Cancer cell expressions of immunoglobulin heavy chains with unique carbohydrate-associated biomarker” Cancer Biomark 5 (4):177-188 (2009). DOI: 10.3233/CBM-2009-0102. PubMed: 19729827. Application: PCR.
- Chaiyakul, M et al. “Cytotoxicity of ORF3 proteins from a nonpathogenic and a pathogenic porcine circovirus” J. Virol. 84 (21):11440-11447 (2010). DOI: 10.1128/JVI.01030-10. PubMed: 20810737. Application: PCR.
- Lee, G et al. “Inhibition of in vitro tumor cell growth by RP215 monoclonal antibody and antibodies raised against its anti-idiotype antibodies” Cancer Immunol. Immunother. 59 (9):1347-1356 (2010). DOI: 10.1007/s00262-010-0864-7. PubMed: 20473495. Application: PCR.
- Bhagwat, B et al. “An in vivo transient expression system can be applied for rapid and effective selection of artificial microRNA constructs for plant stable genetic transformation” J Genet Genomics 40 (5):261-270 (2013). DOI: 10.1016/j.jgg.2013.03.012. PubMed: 23706301. Application: PCR.
- Chaiyakul, M et al. “Cytotoxicity of ORF3 proteins from a nonpathogenic and a pathogenic porcine circovirus” J. Virol. 84 (21):11440-11447 (2010). DOI: 10.1128/JVI.01030-10. PubMed: 20810737. Application: PCR.
- Ahn, CH et al. “Bacterial biofilm-community selection during autohydrogenotrophic reduction of nitrate and perchlorate in ion-exchange brine” Appl. Microbiol. Biotechnol. 81 (6):1169-1177 (2009). DOI: 10.1007/s00253-008-1797-3. PubMed: 19066883. Application: PCR.
- Chi, M et al. “Reduced polyphenol oxidase gene expression and enzymatic browning in potato (Solanum tuberosum L.) with artificial microRNAs” BMC Plant Biol. 14:62 (2014). DOI: 10.1186/1471-2229-14-62. PubMed: 24618103. Application: qRT-PCR.
- Moazzen, H et al. “N-Acetylcysteine prevents congenital heart defects induced by pregestational diabetes” Cardiovasc Diabetol 13:46 (2014). DOI: 10.1186/1475- 2840-13-46. PubMed: 24533448. Application: qRT-PCR.
- Lee, KH et al. “The Histone Demethylase PHF2 Promotes Fat Cell Differentiation as an Epigenetic Activator of Both C/EBPα and C/EBPδ” Mol Cells 37 (10):734-741 (2014). DOI: 10.14348/molcells.2014.0180. PubMed: 25266703. Application: qPCR.
- Romic, S et al. “Does oestradiol attenuate the damaging effects of a fructose-rich diet on cardiac Akt/endothelial nitric oxide synthase signalling?” Br J Nutr 109(11):1940- 1948 (2013). DOI: 10.1017/S0007114512004114. Application: qPCR.
- Chambers, AH et al. “Identification of a strawberry flavor gene candidate using an integrated genetic-genomic-analytical chemistry approach” BMC Genomics 15 (1):217 (2014). DOI: 10.1186/1471-2164-15-217. PubMed: 24742080. Application: qPCR.
- Unsoy, G et al. “Chitosan magnetic nanoparticles for pH responsive Bortezomib release in cancer therapy” Biomedicine & Pharmacotherapy 68(5):641-649 (2014). DOI: 10.1016/j.biopha.2014.04.003. PubMed: 24880680. Application: RT-PCR.
- Kim, SJ et al. “Selective expression of transgene using hypoxia-inducible trans-splicing group I intron ribozyme” J Biotechnol. Pt A 22:7 (2014). DOI: 10.1016/j.jbiotec.2014.10.001. PubMed: 25312327. Application: RT-PCR.
- Li, R et al. “Ultrafine particles from diesel vehicle emissions at different driving cycles induce differential vascular pro-inflammatory responses: implication of chemical components and NF-kappaB signaling” Part Fibre Toxicol 7:6 (2010). DOI: 10.1186/1743-8977-7-6. PubMed: 20307321. Application: RT-PCR.
- Tang, Y et al. “Similar Gene Regulation Patterns for Growth Inhibition of Cancer Cells by RP215 or Anti-Antigen Receptors” J. Cancer Sci. Ther. 5:200-208 (2013). DOI: 10.4172/1948-5956.1000207. Application: RT-PCR.
- Nagarajan, G et al. “Cloning and sequence analysis of IL-2, IL-4 and IFN-γ from Indian Dromedary camels (Camelus dromedarius)” Res. Vet. Sci. 92(3):420-426 (2012). DOI: 10.1016/j.rvsc.2011.03.028. Application: RT-PCR.
- Ker, YB et al. “In vitro polyphenolics erythrocyte model and in vivo chicken embryo model revealed gallic acid to be a potential hemorrhage inducer: physicochemical action mechanisms” Chem. Res. Toxicol 26 (3):325-335 (2013). DOI: 10.1021/tx300456t. Application: PCR Products Viewing.
- Safaee, AG et al. “Disease progression and patient survival are significantly influenced by BRAF protein expression in primary melanoma” Br J Dermatol 169(2):320-8 (2013). DOI: 10.1111/bjd.12351. PubMed: 23550516. Application: PCR Products Viewing.
- Pyo, JS et al. “The prognostic relevance of psammoma bodies and ultrasonographic intratumoral calcifications in papillary thyroid carcinoma” World J Surg 37 (10):2330- 2335 (2013). DOI: 23716027. PubMed: 10.1007/s00268-013-2107-5. Application: PCR Products Viewing.
- Nady, N et al. “Histone recognition by human malignant brain tumor domains” J Mol Biol 423 (5):702-718 (2012). DOI: 10.1016/j.jmb.2012.08.022.. Application: PCR Products Viewing.
- Ish-Shalom, S et al. “Transformation of Botrytis cinerea by direct hyphal blasting or by wound-mediated transformation of sclerotia” BMC Microbiol 11:266 (2011). DOI: 10.1186/1471-2180-11-266. Application: PCR Products Viewing.